|
|


|
The MET-FLAM Faculty
Personal information:
|

|
| Name:
| Peter WOLF
|
| Acad. Degree:
| Univ.-Prof. Dr. med. univ. (MD)
|
| Current Position:
| Chair, Department of Dermatology and Venereology,
Medical University of Graz
|
| Contact Details:
|
Department of Dermatology and Venereology, Medical University of Graz,
Auenbruggerplatz 8, A-8036 Graz;
phone: +43 316 385 80315,
✉ e-mail
|
| Websites:
|
[Department]
[Team]
[Personal]
|
| ORCID:
|
[0000-0001-7777-9444]
|
| Research Metrics:
|
[semanticscholar]
|
Scientific Interests:
The group of Peter Wolf has long-standing experience in translational research in dermatology and focusses on the mechanisms of
inflammatory skin diseases and malignant skin diseases with inflammatory microenvironment in order to develop novel treatment
strategies.
|
Proposed Dissertation Topic:
The role of the metabolome in skin homeostasis and inflammatory skin diseases
Background:
The skin and its appendages and glands produce and store several low molecular-weight compounds (i. e. metabolites)
that either fulfill specific physiological roles or are considered by-products. Skin metabolites originate from sweat, sebum, and protein
degradation in the outer layers of the skin as well as interstitial fluid. Microbes residing in the skin possess a large reservoir of
metabolic enzymes to metabolize molecules, drive metabolic reprogramming of cells, and affect immune responses. In addition, keratinocytes
and skin immune cells metabolize nutrients present in their microenvironment [1]. This might play an important role in
common chronic inflammatory skin diseases such as psoriasis or atopic dermatitis (AD), which are characterized by hyperproliferation of
keratinocytes and an increase in inflammatory cells, suggesting an enhanced nutrient demand of the cells. We have shown that exposomes such
as UV radiation affect the skin microbiome, which in turn can modulate the immune response [2, 3].
Moreover, certain species of the microbiota present in the superficial layers of the skin can metabolize urocanic acid with the cis-isomer
being a key immunosuppressive molecule induced by UV exposure, thereby affecting immune function. Psoriasis, a Th17-type disease, has
a complex pathogenesis involving keratinocytes and immune cells. After successful therapy (by UV radiation or biologic drugs such as
TNFα, IL-17, IL-12/23 or IL-23 inhibitors), psoriasis lesions usually regress completely.
However, after cessation of treatment, the lesions reappear, most often on exactly the same body sites [4].
Hypothesis and objectives:
We hypothesize that metabolic rewiring in AD patients before and after UV exposure and targeted approaches such as blockade of the
IL-4α receptor chain leads to therapeutic success [5]. In psoriasis, we will specifically address
the question of which molecular, cellular, microbial, or metabolic remnants remain in the skin that may reinitiate the inflammatory cycle
and cause psoriasis recurrence. In both diseases, we aim to identify metabolites as potential therapeutic targets to develop new treatment
and prevention strategies [6, 7].
Methods and approaches:
The PhD candidate will learn to investigate skin samples of archived material of patients from clinical studies and biological materials
from mouse models in order to scrutinize the metabolome and identify therapeutic targets. The student will perform flow cytometry, qPCR,
western blotting, multiplex ELISA, immune-bead magnetic cell sorting, multi-color immunohistochemistry / immunofluorescence
microscopy, and immune function assays (T-cell co-proliferation) (1st and 2nd year) and acquire photobiological methods such
as UV light treatment and dosimetry. The student will collaborate with core facilities using LC-MS/MS and NMR to identify metabolites
and exploit the full potential of bioinformatics approaches to gain deep insights into the link between the metabolome and inflammation
(2nd and 3rd year) [8]. Finally, the student will employ in vivo models to validate findings (3rd and 4th year)
[9].
Pitfalls and alternative approaches:
From a technical point of view, the entire methodology required for the execution of the proposed project has been established in the
laboratory of the group, its partners and / or core facilities of the Center for Medical Research of the Medical
University and employed in recent investigations, as outlined in the figure and recent work published [8]. However, we
are aware that the interplay of the microbiota with skin and its cellular elements is highly complex, and therefore it may be challenging
to dissect bystander effects from causal relationships using solely human materials. Alternative work in mouse models and or in ex vivo
human skin models permitting functional testing will help to overcome the caveats in order to scrutinize the metabolome and identify therapeutic
targets.
Involved Faculty members:
Peter Wolf (PI), Vanessa Stadlbauer-Köllner (microbiota analysis),
Gunther Marsche (lipidomics), Ákos Heinemann (cytokines and signaling
pathways), Aitak Farzi (adipocyte function), and Kathrin Eller (immunological assays).
International Collaborations:
Rachael A. Clark (Brigham and Women's Hospital, Harvard Medical School, Boston, USA), Marc Vocanson (Centre International de
Recherche en Infectiologie, Institut National de la Santé et de la Recherche Médicale, U1111, Université Claude
Bernard Lyon 1, Centre National de la Recherche Scientifique, UMR5308, École Normale Supérieure de Lyon,
Université de Lyon, Lyon, France), Karl Quinn (Director Population Health, Metabolon, Research Triangle Park, North
Carolina, USA).
Facilities:
Our team currently consists of two PhD candidates, four postdocs and two technicians. The description of the specific research interests of
the team members is given at the webpage
of the Wolf lab. The laboratories of the group of translational dermatology are located at the Department of Dermatology and in the
Center of Medical Research (ZMF), Medical University of Graz, located across the 2-wing building of the Department of
Dermatology. Equipment for all basic and specific research methods is available in the laboratories of the group. Seahorse technology,
metabolomics and mass spectrometry are available on the campus through collaboration partners.
Preparatory Findings:
The methodology required for the execution of the proposed project has been employed in recent investigations of the group and first data
have been published recently [8].
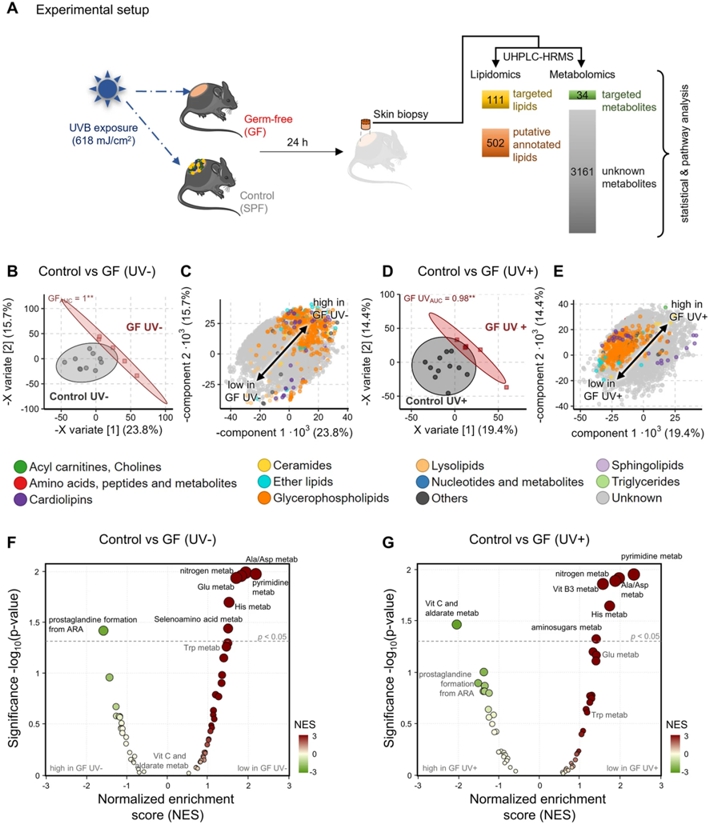
Figure 1. Cutaneous metabolome strongly depends on the presence of microbiome before and after UVB exposure.
(A) Schematic overview of study design. Skin biopsy samples were subjected to metabolomics and lipidomics measurements from
which data was extracted in a targeted and untargeted approach as described in methods.
(B) PLS-DA scores plot investigating the difference of cutaneous metabolome in the absence of any microbiome (GF) before UVB
exposure compared to control mice. Each point represents the metabolome of one mouse skin sample and nearness of points represents metabolic
similarity. ROC analysis with X variate 1– 3 found the metabolomes to differ significantly
(p < 0.001) with an AUC of 1. The 95 % confidence interval of each group is marked by their
coloured ellipse.
(C) Corresponding PLS-DA loadings plot to (B). Each point represents a metabolite’s contribution to the group separation
observed in the scores plot (B) showing that unknown metabolites strongly differ after UVB exposure.
(D) PLS-DA scores plot investigating the difference of cutaneous metabolome in the absence of any microbiome (GF) after UVB
exposure compared to control mice. Points and ellipse as in panel B. ROC analysis with X variate 1– 3
found the metabolomes to differ significantly (p < 0.01) with an AUC of 0.98.
(E) Corresponding PLS-DA loadings plot to (D) [points = metabolites as in (B)] showing that unknown metabolites strongly differ
after UV exposure.
(F) Functional analysis of unknown metabolites in the absence of any microbiome (GF) before UVB exposure compared to control
mice. Only significantly impacted pathways are labelled (p < 0.05).
(G) Functional analysis of unknown metabolites in the absence of any microbiome (GF) after UVB exposure compared to control
mice. Significantly impacted pathways are labelled above the dotted line (p < 0.05).
(n = 6 –10 mice per experimental group; data are pooled from two independent experiments)
(published in [8]).
|
|
References:
-
Patra V, Gallais Sérézal I, Wolf P:
Potential of Skin Microbiome, Pro- and/or Pre-Biotics to Affect Local Cutaneous Responses to UV Exposure.
Nutrients,
2020; 12(6):1795.
![[DOI Journal link] [DOI Journal link]](gifs/doi.gif)
-
Patra V, Wagner K, Arulampalam V, Wolf P:
Skin Microbiome Modulates the Effect of Ultraviolet Radiation on Cellular Response and Immune Function.
iScience,
2019; 15:211–222.
![[DOI Journal link] [DOI Journal link]](gifs/doi.gif)
-
Vieyra-Garcia PA, Wolf P:
A deep dive into UV-based phototherapy: Mechanisms of action and emerging molecular targets in inflammation and cancer.
Pharmacol Ther,
2021; 222:107784.
![[DOI Journal link] [DOI Journal link]](gifs/doi.gif)
-
Benezeder T, Wolf P:
Resolution of plaque-type psoriasis: what is left behind (and reinitiates the disease).
Semin Immunopathol,
2019; 41(6):633–644.
![[DOI Journal link] [DOI Journal link]](gifs/doi.gif)
-
Patra V, Woltsche N, Cerpes U, Bokanovic D, Repelnig, M, Joshi A, Perchthaler I, Fischl M, Vocanson M, Bordag N, Dirdevic M,
Woltsche J, Quehenberger F, Legat F, Wedrich A, Horwath-Winter J, Wolf P:
Persistent neutrophil infiltration and unique ocular surface microbiome typify dupilumab-associated conjunctivitis in patients with atopic dermatitis.
Ophthalmol Sci,
2023, published online before print.
![[DOI Journal link] [DOI Journal link]](gifs/doi.gif)
-
Trakaki A, Wolf P, Weger W, Eichmann TO, Scharnagl H, Stadler JT, Salmhofer W, Knuplez E, Holzer M, Marsche G:
Biological anti-psoriatic therapy profoundly affects high-density lipoprotein function.
Biochim Biophys Acta Mol Cell Biol Lipids,
2021; 1866(7):158943.
![[DOI Journal link] [DOI Journal link]](gifs/doi.gif)
-
Joshi AA, Vocanson M, Nicolas JF, Wolf P, Patra V:
Microbial derived antimicrobial peptides as potential therapeutics in atopic dermatitis.
Front Immunol,
2023; 14:1125635.
![[DOI Journal link] [DOI Journal link]](gifs/doi.gif)
-
Patra V, Bordag N, Clement Y, Köfeler H, Nicolas JF, Vocanson M, Ayciriex S, Wolf P:
Ultraviolet exposure regulates skin metabolome based on the microbiome.
Sci Rep,
2023; 13(1):7207.
![[DOI Journal link] [DOI Journal link]](gifs/doi.gif)
-
Sorger H, Dey S, Vieyra-Garcia PA, Pölöske D, Teufelberger AR, de Araujo ED, Sedighi A, Graf R, Spiegl B, Lazzeri I,
Braun T, Garces de Los Fayos Alonso I, Schlederer M, Timelthaler G, Kodajova P, Pirker C, Surbek M,
Machtinger M, Graier T, Perchthaler I, Pan Y, Fink-Puches R, Cerroni L, Ober J, Otte M, Albrecht JD, Tin G, Abdeldayem A,
Manaswiyoungkul P, Olaoye OO, Metzelder ML, Orlova A, Berger W, Wobser M, Nicolay JP, André F, Nguyen VA, Neubauer HA,
Fleck R, Merkel O, Herling M, Heitzer E, Gunning PT, Kenner L, Moriggl R, Wolf P:
Blocking STAT3/5 through direct or upstream kinase targeting in leukemic cutaneous T-cell lymphoma.
EMBO Mol Med,
2022; 14(12):e15200.
![[DOI Journal link] [DOI Journal link]](gifs/doi.gif)
|
|





![[DOI Journal link] [DOI Journal link]](gifs/doi.gif)